Center for Arthropod Management Technologies (CAMTech)
December 10, 2020
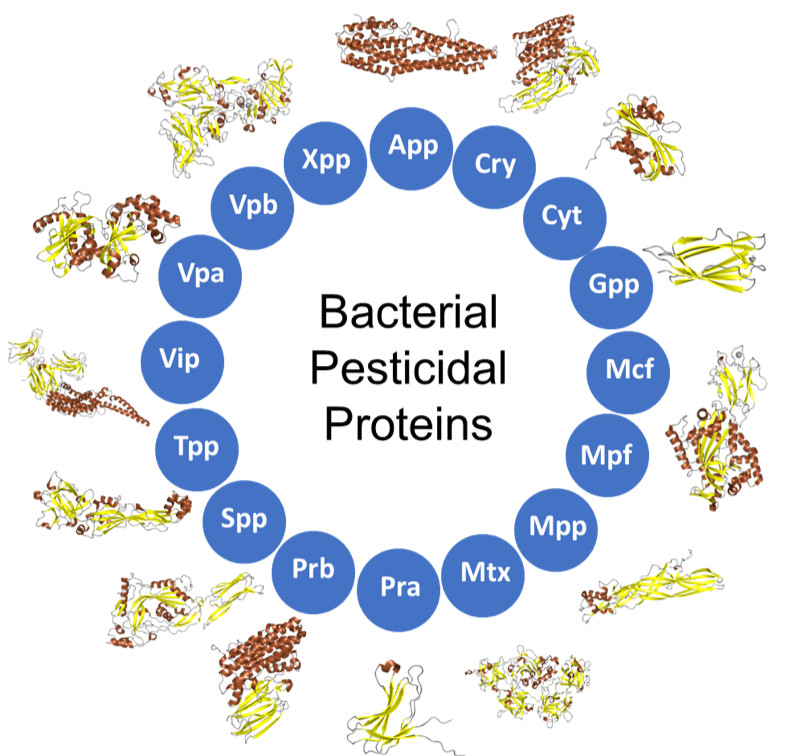
A new way to name certain proteins is a potential gamechanger for researchers developing organic pesticides that target specific insects, which are more environmentally friendly than chemically-based alternatives.
The new naming system—which will replace its predecessor from the 1990s—along with a newly established resource center, will streamline work and increase efficiency in finding and identifying proteins.
A Streamlined System
The naming system is specific to proteins produced by bacteria that are used in pesticides. Today, there are hundreds of these pesticidal proteins—many more than the original naming system was designed to handle. As the number of newly identified proteins continues to rise, a new information resource was needed to help scientists sort through all the proteins and find ones useful to their applications.
Bryony Bonning of the University of Florida, along with colleagues Neil Crickmore of the University of Sussex (who developed the previous system), Colin Berry of Cardiff University, and Suresh Pannerselvam of the University of Florida, designed a new system that describes the structure of the protein in its name. Their work was conducted as part of the Center for Arthropod Management Technologies (CAMTech), an Industry-University Cooperative Research Center (IUCRC) funded by the National Science Foundation.
Proteins have tremendous variation in their structure. Since their structure is related to their function, the shape of pesticidal proteins can predict what insects they might act against or whether they might interact with other proteins. The research team used this relationship between structure and function in the protein’s name by developing a system that puts pesticidal proteins into groups distinguished by shape. With the new system, researchers can infer what type of pesticide might be made with a protein just based on its name.
“It will really help the field,” said one of the project leads, Bryony Bonning, a researcher at the University of Florida whose research is funded by the IUCRC program. “It will speed things up, particularly with the immense number of pesticidal proteins being identified now with advanced gene sequencing technologies.”
Researchers around the world study different strains of Bacillus thuringiensis and other pesticidal-producing bacteria in order to identify new proteins that can be used to create improved pesticides. This work also helps researchers bioengineer plants to produce their own bacteria-derived pesticidal proteins that protect the crops from pests.
To ensure the new naming system would be beneficial to a wide range of users, it was developed in collaboration with industry partners and has also been presented to government agencies like the Environmental Protection Agency, the U.S. Food and Drug Administration and the USDA's Animal and Plant Health Inspection Service.
“We didn’t come up with this system alone,” Bonning said. “Neil presented the idea multiple times at different meetings and got feedback from industry representatives and other academics. It was definitely an iterative process with the naming system refined with input from industry.”

New Resource Center
The new pesticidal protein naming system now has its own online database. Called the Bacterial Pesticidal Protein Resource Center, researchers can access the website to look up proteins, cross-reference old names with new ones, and submit new proteins to be named. To date, there are over 1,000 pesticidal proteins in the system.
“The extraordinary number and diversity of pesticidal proteins coming from microbial genomes and metagenomes was overwhelming our capacity to deal with them,” said collaborator Jim Baum, a researcher at Bayer. The sheer number of proteins made it hard for the industry to work with, so they strongly supported funding this project to create a more usable system.
Along with old and new names, the database also contains information on protein structures for those that are publicly available. For newly discovered proteins that industry wants to remain secret for patented use, the database also has a private submission option. The database and protein analysis tools can also be downloaded from the site for remote use.
Ultimately, the new system will increase efficiency by making it easier to swiftly identify proteins of particular interest. The resource center will increase awareness of the diversity of these pesticidal proteins and help researchers share their knowledge. The system has also been largely automated to speed up and ease the naming process as new proteins are being discovered at an increasing pace.
